Recently, The clinical results of Molnupiravir (MK-4482, EIDD-2801), a novel coronavirus small molecule developed by Merk, are relatively exciting. Here is a focus for a brief look at its bio-synergy and complex structure. At the meanwhile, the mechanism and structure of Remdesivir (considered as people’s hope in last year) are also mentioned.

Molnupiravir is a prodrug that is transformed in vivo with triphosphate (see picture 2)
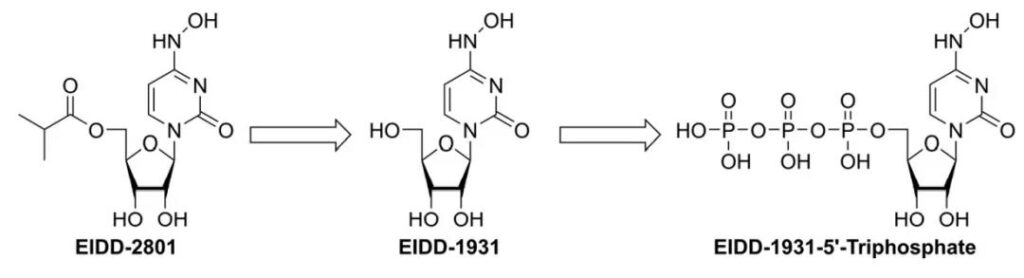
Let’s get down to business
Molnupiravir inhibits many RNA viruses, including the novel coronavirus, by introducing a large number of mutations in the virus’s RNA that “paralyzes” the virus’s genome, rendering it unable to perform its task properly due to genetic errors.
In picture3, when activated Molnupiravir (M in orange color) presents, RdRp (RNA-dependent RNA polymerase, simply known as RNA replicase) mistakenly adds it to —gRNA. The +gRNA (viral genome) is then generated using the -grNA that has infiltrated M as A template, which introduces A number of errors, including infiltration of M and base “A”/” G “transitions. In other words, a large number of mutations were introduced into the virus genome, and the genetic information of the virus was “tampered with” so much that it almost completely lost its normal function.

Why does Molnupiravir introduce A mutation into viral RNA (causing A/G conversion)? It starts with its chemical structure.Because it has two main tautomers, called imino-m and Amino-M respectively. As shown in Figure 4, imino-M on the left is complementary paired with base A to form two HB, while Amino-M on the right is complementary paired with base G to form three HB.
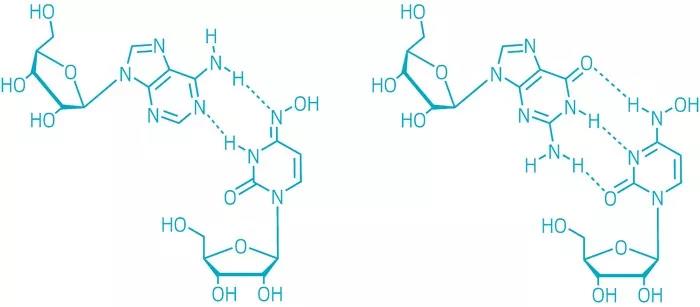
It is well known that DNA or RNA (other than chemical bonding) relies primarily on two forces: base stacking and HB pairing, with A normally pairing with U and G with C. Molnupiravir’s different tautomers, which mimic U or C, can pair with A or G, respectively.
The complex structure of two tautomers of “M” paired with A or G, PDB ID 7OZU & 7OZV, cryo-electron microscopy structure, resolution 3.3A & 3.2A has been resolved. See picture 5

Unlike Molnupiravir, which introduces A/G swap (mutation) into viral RNA through A different tautomer, Remdesivir is A nucleoside analogue (and prodrug) that also acts on RdRp, but the mechanism is different.

Remdesivir also carries triphosphate in the body, and its structure is similar to ATP except for a slight difference in the N of the base, mainly due to an extra CN (upper left corner of Figure 7).
It is this CN that, when RTP (activated Remdesivir) fish passes into the RNA polymerase, “jams” the assembly line due to steric hindrance, causing RNA synthesis to fail and RdRp to fail.
As shown in Figure 7E/F/G/H, the normal base (gray) and Ser861 (blue and purple) did not clash and passed normally, while RTP (orange) was “stuck” and could not pass due to the extra CN, thus obstructing the replication of viral RNA.

In general, Remdesivir seems to have limited efficacy and does not really serve as the “people’s hope,” but Molnupiravir has recently shown promise and is oral and has an advantage over Remdesivir (IV).
Both are nucleoside analogues with the same target, but the specific mechanism is different. Molnupiravir appears to be better than Remdesivir at present, but more time is needed. Just like our old saying:”It takes three days to burn the jade, and seven years to distinguish the material.”
- References:
- Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis
- Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication
- An emerging antiviral takes aim at COVID-19



